Research Highlights
Gene discovery in autism spectrum disorder and other developmental disorders
Exome sequencing has revolutionized our ability to find the genes associated with autism spectrum disorder (ASD) and other severe developmental disorders. Four papers, published concurrently in 2012, showed an excess of de novo mutations that disrupt genes in cases compared to controls and Dr. Sanders, working in Dr. State’s lab at Yale, was the first author of one of these (Sanders et al. Nature 2012). This paper also demonstrated that observing disruptive de novo mutations in the same gene across multiple cases could be used to establish genome-wide association with ASD, and this method was used to identify the voltage-gated sodium channel SCN2A as one of the most frequent causes of ASD. As a member of the Autism Sequencing Consortium, the Sanders Lab continues to play an active role in advancing gene discovery in ASD. The initial observation of an excess of disruptive de novo mutations in cases has been replicated in larger cohorts (e.g. De Rubeis et al. Nature 2014, Iossifov et al. Nature 2014, Dong et al. 2014). These two cohorts were combined and integrated with copy number variant (CNV) data to identify 65 genes associated with ASD (Sanders et al. Neuron 2015). This approach to gene discovery has been successful in several other developmental disorders, including severe developmental delay, epileptic encephalopathy, and congenital heart disease.
Understanding the role of ASD genes
The genes discovered through exome sequencing provide an entree into the biology of ASD, hopefully as a foundation for developing theraputics. The Sanders Lab takes two research approaches: 1) To consider ASD-associated genes en masse to look for patterns of convergence State MW, Šestan N Science 2012; 2) To investigate the genes with the strongest evidence of ASD association, such as SCN2A in depth.
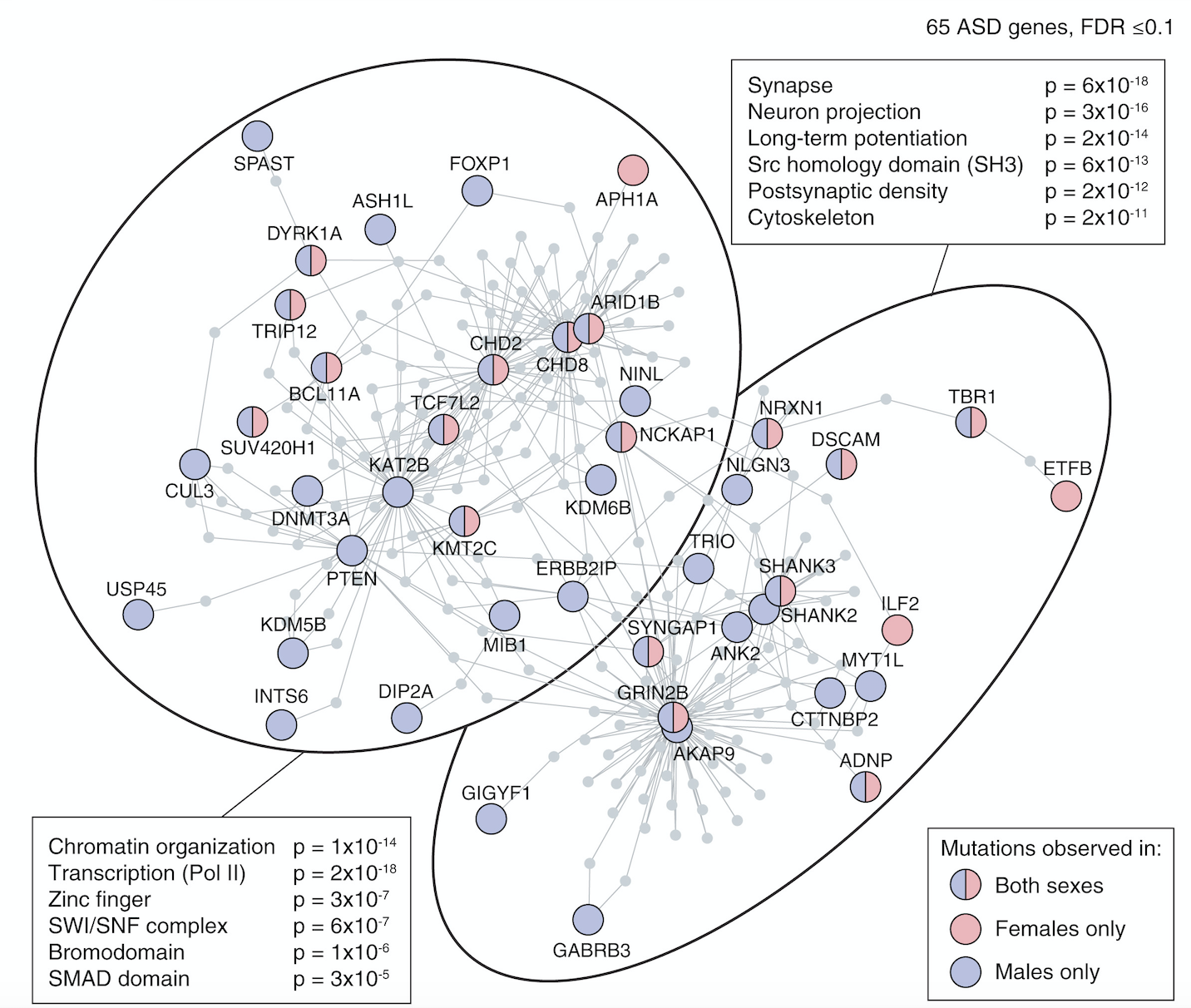
Through the first approach, the Sanders Lab has helped delineate two major functional roles of ASD-associated genes: 1) gene expression regulators (GER), i.e. genes that influence the expression of other genes such as CHD8 and ARID1B; and 2) neuronal communication, such as SCN2A and NRXN1, see De Rubeis et al. Nature 2014 and Sanders et al. Neuron 2015. Co-expression analysis showed convergence across multiple ASD-associated genes during mid-fetal development in the prefrontal cortex and this result was driven by layer 5/6 glutamatergic neurons (Willsey et al. Cell 2015). Similar patterns have been identified through other systems-based approaches, reviewed in Sanders Curr Opin Genet Dev 2015.
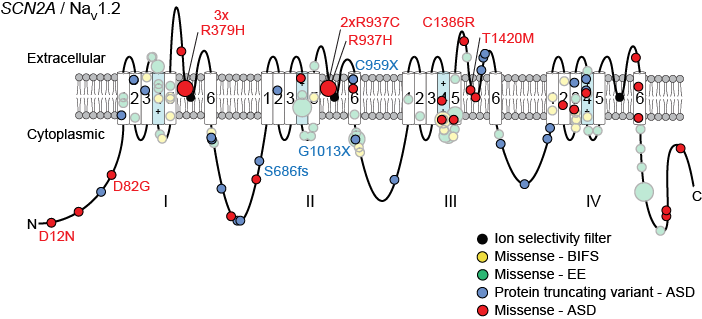
The second approach has focused mainly on the gene SCN2A and is performed in collaboration with the Bender lab at UCSF, the SCN2A family group, and Simons VIP. By integrating genetic data across multiple cases and cohorts with structural data on the NaV1.2 protein encoded by SCN2A and electrophysiology, we demonstrated that loss-of-function mutations of SCN2A lead to ASD and developmental delay, while gain-of-function mutations lead to epileptic encephalopathy (Ben-Shalom et al. 2017).
Copy number variants (CNVs) in autism spectrum
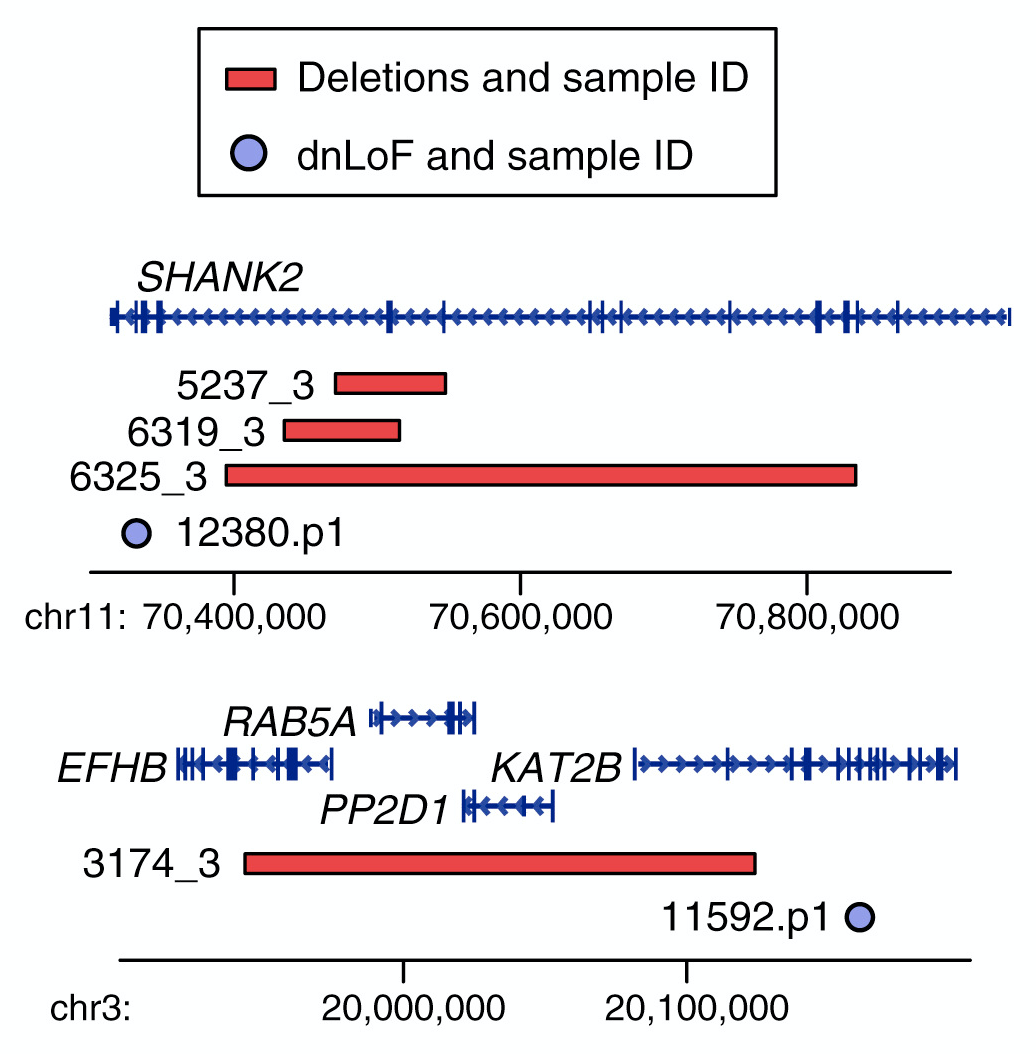
My early publications describe the development of a novel approach to detecting de novo CNVs in Illumina SNP genotyping data and applying this to Autism Spectrum Disorder (ASD) families from the Simons Simplex Collection (SSC). While de novo CNVs had previously been associated with ASD (Sebat et al. 2007), our analysis of this cohort of 1,124 families, including unaffected siblings, allowed us to clarify the genomic architecture. In turn, this allowed us to develop a statistical approach to evaluating whether specific genomic loci are associated with ASD; four de novo duplications at 7q11.23 met these criteria for ASD association with reduced sociability and, notably, deletions at the same locus are associated with increased social behavior in William’s Syndrome. We have recently (2015) published a CNV analysis of all 2,591 families in the SSC. By integrating CNV and exome data we showed that small de novo deletions contain one critical ASD gene, while large de novo CNVs contain multiple ASD risk genes. We use this finding to identify six ASD risk CNV loci and 65 ASD risk genes. I led the analysis and write up of both projects including the development of novel approaches to CNV detection. Autism Speaks cited our 2011 CNV analysis as one of the top Ten Autism Research Achievements.